Engineering scarless chromosomal rearrangements using homology-mediated recombination and selection
Genetically engineered mouse models of human cancers have proven to be essential for the elucidation of basic molecular mechanisms of cancer pathogenesis and their therapeutic targeting. However, for many genetic aberrations, such as those recently identified in therapy resistant leukemias, conventional modeling approaches are either not feasible or entirely impossible. For example, childhood leukemias with t(8;16) chromosomal rearrangements are associated with resistance to conventional chemotherapy and result in the expression of the MOZ-CBP fusion oncoprotein. The MOZ-CBP fusion oncoprotein is over 400 kDa in size, which is too large to be expressed as a retroviral cDNA transgene. Given the laborious requirements for conventional engineering of chromosomal translocations in mice, no faithful preclinical models of t(8;16) leukemias exist. Similarly, chemotherapy-resistant NUP98 fusion oncoproteins, such as NUP98-NSD1 from t(5;11) and NUP98-JARID1A from t(11;12) chromosomal translocations, cannot be accurately modeled by cDNA transgenic expression due to the artifacts of retroviral overexpression and uncertainty about the relevant cell of origin of these leukemias. Lastly, even for relatively simple chromosomal rearrangements such as inv(3) associated with poor prognosis and therapy resistance in AML, current models of EVI1 overexpression are not accurate, because this rearrangement causes at least 3 simultaneous molecular aberrations: overexpression of EVI1, as well as dysregulation of RPN1 and GATA2 which flank the leukemic chromosomal inversion. Thus, the field lacks accurate preclinical models for virtually all of the chromosomal rearrangements and fusion oncogenes that subsume normal developmental and gene regulatory programs in refractory childhood leukemias.
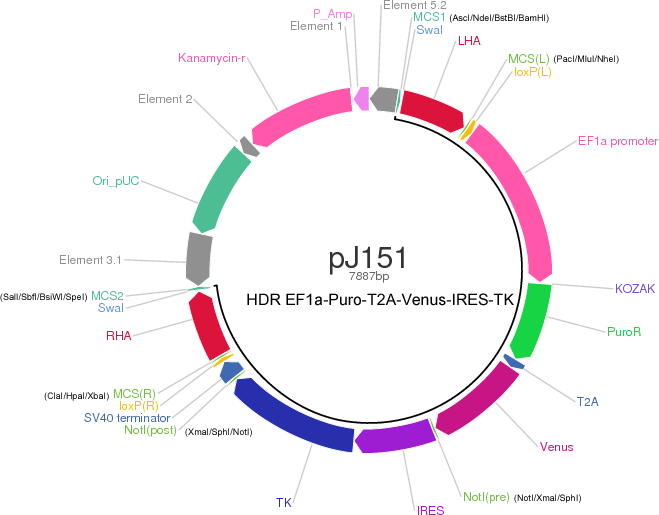
Recently, CRISPR technology has been used for somatic engineering of oncogenic chromosomal rearrangements. To facilitate the engineering of chromosomal rearrangements that may be induced at relatively low frequency by CRISPR, we have developed a homology-directed repair (HDR) vector. This vector, available openly from Addgene, contains two recombination homology arms and loxP-flanked cassette encoding puromycin resistance gene, Venus fluorescence marker, thymidine kinase suicide gene, for scarless chromosomal engineering. The recombination homology arms can be easily replaced by restriction enzyme cloning, and the resultant vector can be delivered into cells by electroporation. Consequently, the cells can be selected by FACS and/or puromycin treatment, and the resultant rearrangements can be confirmed. Upon confirmation of the intended rearrangement, the cells can then be transfected or treated with the Cre recombinase to excise the targeting cassette, and cells with complete excision can be isolated by ganciclovir treatment.